Technology
Nuventure Connect Launches AI Innovation Lab for Smart Industrial Innovation
The company also unveiled NuWave, an IoT platform under development that integrates AI-driven automation, real-time analytics, and predictive maintenance tools

Kerala-based technology firm Nuventure Connect has launched an AI Innovation Lab aimed at accelerating the development of intelligent, sustainable industrial solutions. The launch coincides with the company’s 15th anniversary and signals a renewed focus on artificial intelligence (AI), Internet of Things (IoT), and data-driven automation.
The new lab will serve as a collaborative platform for enterprises, startups, and small-to-medium-sized businesses to design, test, and refine next-generation technologies. Nuventure, which specializes in deep-tech solutions and digital transformation, will provide technical expertise in AI, machine learning, and IoT to support co-development efforts.
“Our vision is to help businesses harness the full potential of AI and IoT to optimize operations and improve sustainability,” said Tinu Cleatus, Managing Director and CEO of Nuventure Connect.
The company also unveiled NuWave, an IoT platform under development that integrates AI-driven automation, real-time analytics, and predictive maintenance tools. The platform is targeted at industries seeking to cut energy consumption and preempt operational failures. Company representatives showcased solutions that could potentially reduce industrial energy costs by up to 40%.
Nuventure is inviting organizations to partner with the lab through structured collaboration models. Participating firms will gain access to advanced IoT infrastructure, expert mentorship, and opportunities to co-create pilot projects.
The initiative places Nuventure among a growing number of regional tech firms contributing to global trends in sustainable and AI-led industrial innovation. By opening its lab to cross-sector partnerships, the company aims to help shape the next phase of digital transformation in manufacturing and beyond.
Society
How 2025’s Emerging Technologies Could Redefine Our Lives

In an age when algorithms help cars avoid traffic and synthetic microbes could soon deliver our medicine, the boundary between science fiction and science fact is shrinking. The World Economic Forum’s Top 10 Emerging Technologies of 2025 offers a powerful reminder that innovation is not just accelerating — it’s converging, maturing, and aligning itself to confront humanity’s most urgent challenges.
From smart cities to sustainable farming, from cutting-edge therapeutics to low-impact energy, this year’s list is more than a forecast. It’s a blueprint for a near future in which resilience and responsibility are just as crucial as raw invention.
Sensing the World Together
Imagine a city that can sense a traffic jam, redirect ambulances instantly, or coordinate drone deliveries without a hiccup. That’s the promise of collaborative sensing, a leading entry in the 2025 lineup. This technology enables vehicles, emergency services, and infrastructure to “talk” to each other in real time using a network of connected sensors — helping cities become safer, faster, and more responsive.
It’s one of several technologies on this year’s list that fall under the theme of “trust and safety in a connected world” — a trend reflecting the growing importance of reliable information, responsive systems, and secure networks in daily life.
Trust, Truth, and Invisible Watermarks
But as digital content spreads and AI-generated images become harder to distinguish from reality, how do we safeguard truth? Generative watermarking offers a promising solution. By embedding invisible tags in AI-generated media, this technology makes it easier to verify content authenticity, helping fight misinformation and deepfakes.
“The path from breakthrough research to tangible societal progress depends on transparency, collaboration, and open science,” said Frederick Fenter, Chief Executive Editor of Frontiers, in a media statement issued alongside the report. “Together with the World Economic Forum, we have once again delivered trusted, evidence-based insights on emerging technologies that will shape a better future for all.”
Rethinking Industry, Naturally
Other breakthroughs are tackling the environmental consequences of how we make things.
Green nitrogen fixation, for instance, offers a cleaner way to produce fertilizers — traditionally one of agriculture’s biggest polluters. By using electricity instead of fossil fuels to bind nitrogen, this method could slash emissions while helping feed a growing planet.
Then there’s nanozymes — synthetic materials that mimic enzymes but are more stable, affordable, and versatile. Their potential applications range from improving diagnostics to cleaning up industrial waste, marking a shift toward smarter, greener manufacturing.
These technologies fall under the trend the report identifies as “sustainable industry redesign.”
Health Breakthroughs, From Microbes to Molecules
The 2025 report also spotlights next-generation biotechnologies for health, a category that includes some of the most exciting and potentially transformative innovations.
Engineered living therapeutics — beneficial bacteria genetically modified to detect and treat disease from within the body — could make chronic care both cheaper and more effective.
Meanwhile, GLP-1 agonists, drugs first developed for diabetes and obesity, are now showing promise in treating Alzheimer’s and Parkinson’s — diseases for which few options exist.
And with autonomous biochemical sensing, tiny wireless devices capable of monitoring environmental or health conditions 24/7 could allow early detection of pollution or disease — offering critical tools in a world facing climate stress and health inequities.
Building Smarter, Powering Cleaner
Under the theme of “energy and material integration”, the report also identifies new approaches to building and powering the future.
Structural battery composites, for example, are materials that can both carry loads and store energy. Used in vehicles and aircraft, they could lighten the load — quite literally — for electric transportation.
Osmotic power systems offer another intriguing frontier: by harnessing the energy released when freshwater and saltwater mix, they provide a low-impact, consistent power source suited to estuaries and coastal areas.
And as global electricity demand climbs — especially with the growth of AI, data centers, and electrification — advanced nuclear technologies are gaining renewed interest. With smaller, safer designs and new cooling systems, next-gen nuclear promises to deliver scalable zero-carbon power.
Toward a Converging Future
This year’s edition of the report emphasizes a deeper trend: technological convergence. Across domains, innovations are beginning to merge — batteries into structures, biology into computing, sensing into infrastructure. The future, it seems, will be shaped less by standalone inventions and more by integrated, systemic solutions.
“Scientific and technological breakthroughs are advancing rapidly, even as the global environment for innovation grows more complex,” said Jeremy Jurgens, Managing Director of the World Economic Forum, in the WEF’s official media release.
“The research provides top global leaders with a clear view of which technologies are approaching readiness, how they could solve the world’s pressing problems and what’s required to bring them to scale responsibly,” he added.
Beyond the Hype
Now in its 13th year, the Top 10 Emerging Technologies report has a strong track record of identifying breakthroughs poised to move from lab to life — including mRNA vaccines, flexible batteries, and CRISPR-based gene editing.
But this year’s list is not just a celebration of possibility. It’s a reminder of what’s needed to deliver impact at scale: responsible governance, sustained investment, and public trust.
As Jeremy Jurgens noted, “Breakthroughs must be supported by the right environment — transparent, collaborative, and scalable — if they are to benefit society at large.”
In a time of climate stress, digital overload, and health inequity, these ten technologies offer something rare: a credible roadmap to a better future — not decades away, but just around the corner.
Space & Physics
MIT unveils an ultra-efficient 5G receiver that may supercharge future smart devices
A key innovation lies in the chip’s clever use of a phenomenon called the Miller effect, which allows small capacitors to perform like larger ones

A team of MIT researchers has developed a groundbreaking wireless receiver that could transform the future of Internet of Things (IoT) devices by dramatically improving energy efficiency and resilience to signal interference.
Designed for use in compact, battery-powered smart gadgets—like health monitors, environmental sensors, and industrial trackers—the new chip consumes less than a milliwatt of power and is roughly 30 times more resistant to certain types of interference than conventional receivers.
“This receiver could help expand the capabilities of IoT gadgets,” said Soroush Araei, an electrical engineering graduate student at MIT and lead author of the study, in a media statement. “Devices could become smaller, last longer on a battery, and work more reliably in crowded wireless environments like factory floors or smart cities.”
The chip, recently unveiled at the IEEE Radio Frequency Integrated Circuits Symposium, stands out for its novel use of passive filtering and ultra-small capacitors controlled by tiny switches. These switches require far less power than those typically found in existing IoT receivers.
A key innovation lies in the chip’s clever use of a phenomenon called the Miller effect, which allows small capacitors to perform like larger ones. This means the receiver achieves necessary filtering without relying on bulky components, keeping the circuit size under 0.05 square millimeters.
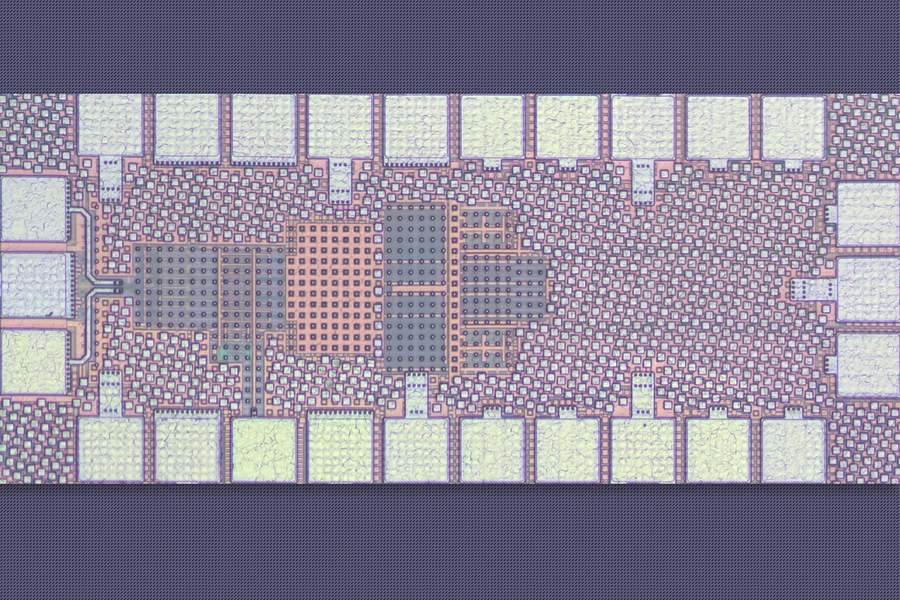
Traditional IoT receivers rely on fixed-frequency filters to block interference, but next-generation 5G-compatible devices need to operate across wider frequency ranges. The MIT design meets this demand using an innovative on-chip switch-capacitor network that blocks unwanted harmonic interference early in the signal chain—before it gets amplified and digitized.
Another critical breakthrough is a technique called bootstrap clocking, which ensures the miniature switches operate correctly even at a low power supply of just 0.6 volts. This helps maintain reliability without adding complex circuitry or draining battery life.
The chip’s minimalist design—using fewer and smaller components—also reduces signal leakage and manufacturing costs, making it well-suited for mass production.
Looking ahead, the MIT team is exploring ways to run the receiver without any dedicated power source—possibly by harvesting ambient energy from nearby Wi-Fi or Bluetooth signals.
The research was conducted by Araei alongside Mohammad Barzgari, Haibo Yang, and senior author Professor Negar Reiskarimian of MIT’s Microsystems Technology Laboratories.
Society
Researchers Unveil Light-Speed AI Chip to Power Next-Gen Wireless and Edge Devices
This could transform the future of wireless communication and edge computing

In a breakthrough that could transform the future of wireless communication and edge computing, engineers at MIT have developed a novel AI hardware accelerator capable of processing wireless signals at the speed of light. The new optical chip, built for signal classification, achieves nanosecond-level performance—up to 100 times faster than conventional digital processors—while consuming dramatically less energy.
With wireless spectrum under growing strain from billions of connected devices, from teleworking laptops to smart sensors, managing bandwidth has become a critical challenge. Artificial intelligence offers a path forward, but most existing AI models are too slow and power-hungry to operate in real time on wireless devices.
The MIT solution, known as MAFT-ONN (Multiplicative Analog Frequency Transform Optical Neural Network), could be a game-changer.
“There are many applications that would be enabled by edge devices that are capable of analyzing wireless signals,” said Prof. Dirk Englund, senior author of the study, in a media statement. “What we’ve presented in our paper could open up many possibilities for real-time and reliable AI inference. This work is the beginning of something that could be quite impactful.”
Published in Science Advances, the research describes how MAFT-ONN classifies signals in just 120 nanoseconds, using a compact optical chip that performs deep-learning tasks using light rather than electricity. Unlike traditional systems that convert signals to images before processing, the MIT design processes raw wireless data directly in the frequency domain—eliminating delays and reducing energy usage.
“We can fit 10,000 neurons onto a single device and compute the necessary multiplications in a single shot,” said Ronald Davis III, lead author and recent MIT PhD graduate.
The device achieved over 85% accuracy in a single shot, and with multiple measurements, it converges to above 99% accuracy, making it both fast and reliable.
Beyond wireless communications, the technology holds promise for edge AI in autonomous vehicles, smart medical devices, and future 6G networks, where real-time response is critical. By embedding ultra-fast AI directly into devices, this innovation could help cars react to hazards instantly or allow pacemakers to adapt to a patient’s heart rhythm in real-time.
Future work will focus on scaling the chip with multiplexing schemes and expanding its ability to handle more complex AI tasks, including transformer models and large language models (LLMs).
-

 Society4 months ago
Society4 months agoStarliner crew challenge rhetoric, says they were never “stranded”
-

 Space & Physics3 months ago
Space & Physics3 months agoCould dark energy be a trick played by time?
-

 Earth4 months ago
Earth4 months agoHow IIT Kanpur is Paving the Way for a Solar-Powered Future in India’s Energy Transition
-

 Space & Physics3 months ago
Space & Physics3 months agoSunita Williams aged less in space due to time dilation
-

 Learning & Teaching4 months ago
Learning & Teaching4 months agoCanine Cognitive Abilities: Memory, Intelligence, and Human Interaction
-

 Earth2 months ago
Earth2 months ago122 Forests, 3.2 Million Trees: How One Man Built the World’s Largest Miyawaki Forest
-

 Women In Science3 months ago
Women In Science3 months agoNeena Gupta: Shaping the Future of Algebraic Geometry
-

 Society5 months ago
Society5 months agoSustainable Farming: The Microgreens Model from Kerala, South India












